
|
Enter a Search String | | Special character and space not allowed in the query term.
Search string should be at least 2 characters long. |
Parameters for LacZ (Pathway Number 141) Pathway Layout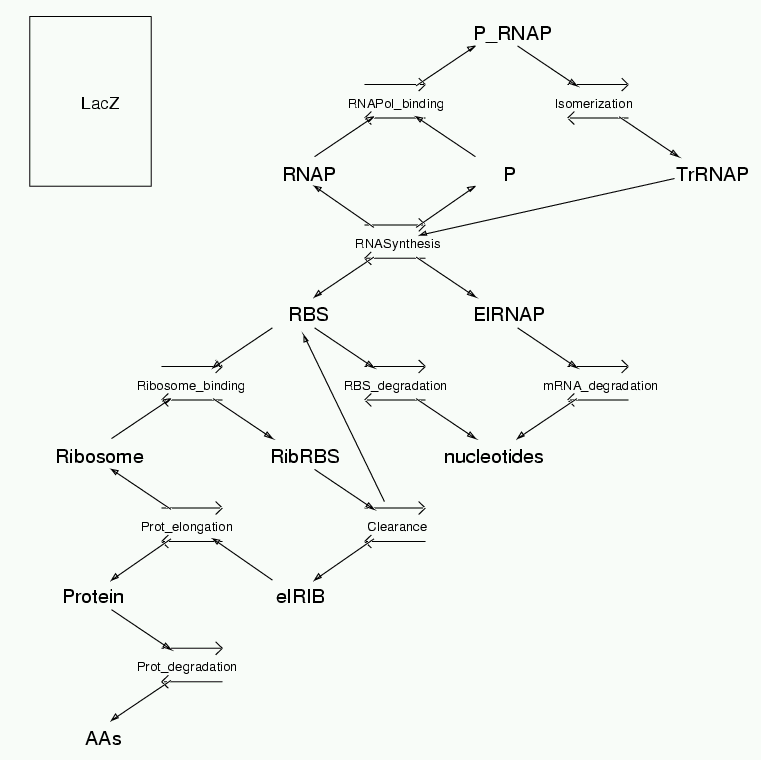
Pathway Basic Parameters | Pathway Name | LacZ | | Pathway Entry Number | 141 | | Accession Number | 29 | | Accession Name | Kierzek_LacZ | | Species | E. Coli | | Tissue | - | | CellCompartment | Internal | | Related Pathways |
| | Notes | Based on Kierzek, Zaim and Zielenkiewicz 2001 JBC 276(11):8165-8172 This is NOT an exact duplicate of their model. Simulation results come out about 1/3 of their values. There are two main discrepancies: they use stochastic simulations, and they also assume linear growth of the cell volume throughout the simulation. The first approximation has been tested using the stochastic simulation mode in Kinetikit, and the differences are small. The second approximation is problematic. By starting the cell at 0.5e-18 m^3, running it for half its cycle, then promoting it to full size of 1e-18 m^3 and finishing off the cycle one sees a closer match to their results for protein levels. Anyway, the purpose of this implementation is to provide a reference to the original simulation.
| Statistics Conversion formatThis pathway is part of accession 29 and is completely specified in the file acc29.g.
There is no separate files for just this pathway. |
Pathway Detail Molecule List Enzyme List Reaction List
| Database compilation and code copyright (C) 2022, Upinder S. Bhalla and NCBS/TIFR
This Copyright is applied to ensure that the contents of this database remain freely available. Please see FAQ for details. |
|

