
 |  NOS_Phosph_ NOS_Phosph_
regulation |
|
Enter a Search String | | Special character and space not allowed in the query term.
Search string should be at least 2 characters long. |
Accession information for NOS_Phosph_regulation (Accession Number 20)
Reaction Scheme
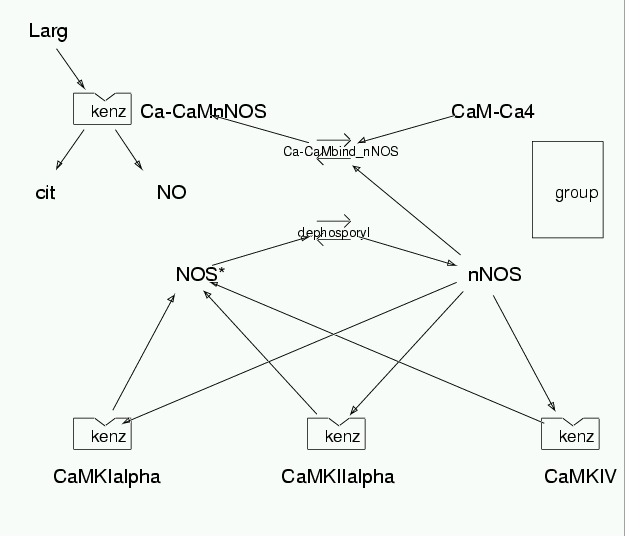
Accession Basic Parameters
| Name | NOS_Phosph_regulation |
| Accession Type | Pathway |
| Transcriber | Sudhir Sivakumaran, NCBS |
| Developer | Sudhir Sivakumaran, NCBS |
| Entry Date (YYYY-MM-DD) | 2001-12-25 00:00:00 |
| Species | rat; Mammalian |
| Tissue | Brain - Neuronal; expressed in E.coli |
| Cell Compartment | Cytosol |
| Source | In-house |
| Methodology | Quantitative match to experiments, Qualitative |
| Model Implementation | Exact GENESIS implementation |
| Model Validation | Approximates orginal data, Quantitatively predicts new data |
| Notes | This model features the phosphorylation of rat brain neuronal NOS expressed in E. coli or Sf9 cells, which leads to a decrease in Vmax of the phosphorylated enzyme, with little change of both the Km for L-arginine and Kact for CaM. This is based on Hayashi Y. et al. J Biol Chem. (1999) 274(29):20597-602. They report of phosphorylatin being carried out by CaM kinases I alpha, II alpha and IV. The rates used have been obtained from their paper and from other reported experimental data. |
Conversion formats
| Database compilation and code copyright (C) 2022, Upinder S. Bhalla and NCBS/TIFR
This Copyright is applied to ensure that the contents of this database remain freely available. Please see FAQ for details. |
|

