
 |  Chemotaxis Chemotaxis |
|
Enter a Search String | | Special character and space not allowed in the query term.
Search string should be at least 2 characters long. |
Accession information for Chemotaxis (Accession Number 55)
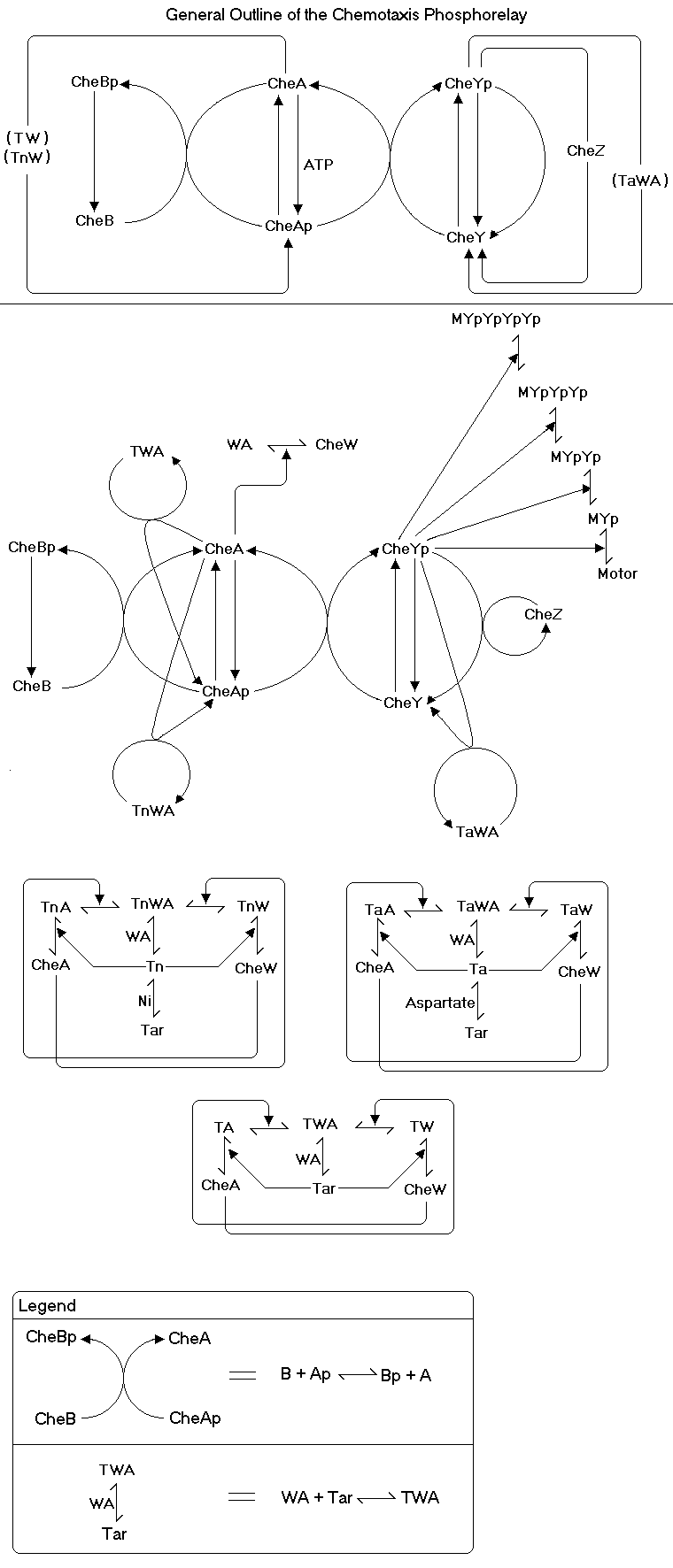
Reaction Scheme
Accession Basic Parameters
| Name | Chemotaxis |
| Accession Type | Pathway |
| Transcriber | Sharat J. Vayttaden, NCBS |
| Developer | Dennis Bray, Robert B. Bourret and, Melvin I. Simon, Cambridge and Caltech |
| Entry Date (YYYY-MM-DD) | 2003-06-10 00:00:00 |
| Species | E. Coli |
| Tissue | - |
| Cell Compartment | Cell membrane + cytosol |
| Source | Bray et al. Mol.Biol.Cell (1993) 4(5): 469-482. ( peer-reviewed publication ) |
| Methodology | Quantitative match to experiments |
| Model Implementation | Mathematically equivalent |
| Model Validation | Approximates original data |
| Notes | All parameters used are from the .BCT files for BCT1.1 provided by Matthew Levin from the Computational Biology Group in the Department of Zoology at the University of Cambridge.Bias does not reach 0.7, there is a lag in the response of bias to the 5 sec 1 uM Aspartate pulse as shown by Bray et al. Mol.Biol.Cell (1993) 4(5): 469-482. The June 2003 version of the BCT program is BCT4.3 and is available at the computational biology site of the Zoology department at Cambridge University. |
Conversion formats| Format | File | | Native Format (GENESIS format) | acc55.g | | GENESIS Format (Annotated version) | Anno_acc55.g |
| Database compilation and code copyright (C) 2022, Upinder S. Bhalla and NCBS/TIFR
This Copyright is applied to ensure that the contents of this database remain freely available. Please see FAQ for details. |
|

