
 |  Jak-Stat_ Jak-Stat_
Pathway |
|
Enter a Search String | | Special character and space not allowed in the query term.
Search string should be at least 2 characters long. |
Accession information for Jak-Stat_Pathway (Accession Number 66)
Reaction Scheme
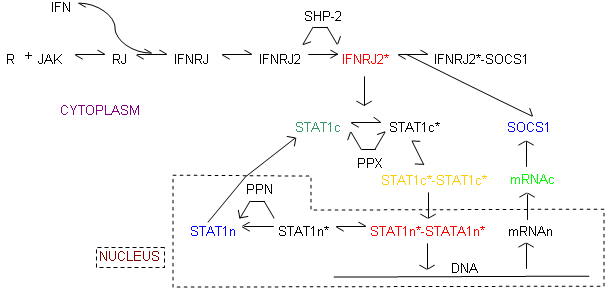
Accession Basic Parameters
| Name | Jak-Stat_Pathway |
| Accession Type | Pathway |
| Transcriber | Sharat J. Vayttaden and Prasoon Agarwal, NCBS |
| Developer | Yamada S, Shiono S, Joo A, Yoshimura A |
| Entry Date (YYYY-MM-DD) | 2006-02-13 00:00:00 |
| Species | mouse |
| Tissue | Liver |
| Cell Compartment | Cytosol, Nucleus |
| Source | Yamada S et al. FEBS Letters 2003 Jan 16;534(1-3):190-6. ( Peer-reviewed publication ) |
| Methodology | Quantitative match to experiments |
| Model Implementation | Exact GENESIS implementation |
| Model Validation | Replicates original data |
| Notes | This model was taken from the Yamada S et al. FEBS Letters 2003 Jan 16;534(1-3):190-6
This model shows the control mechanism of Jak-Stat pathway, here SOCS1 (Suppressor of cytokine signaling-I) was identified as the negative regulator of Jak and STAT signal transduction pathway.
Note: There are a few ambiguities in the paper like initial concentration of IFN and some reactions were missing in the paper that were employed for obtaining the results. The graphs are almost similar to the graphs as shown in the paper but still some ambiguities regarding the concentration are there. Thanks to Dr Satoshi Yamada for clarifying some of those ambiguities and providing the values used in the simulations. |
Conversion formats
| Database compilation and code copyright (C) 2022, Upinder S. Bhalla and NCBS/TIFR
This Copyright is applied to ensure that the contents of this database remain freely available. Please see FAQ for details. |
|

