 |  |  GC GC |
Enter a Search String | | Special character and space not allowed in the query term.
Search string should be at least 2 characters long. |
Parameters for Shared_Object_cGMP_regulation (Pathway Number 176) Pathway Layout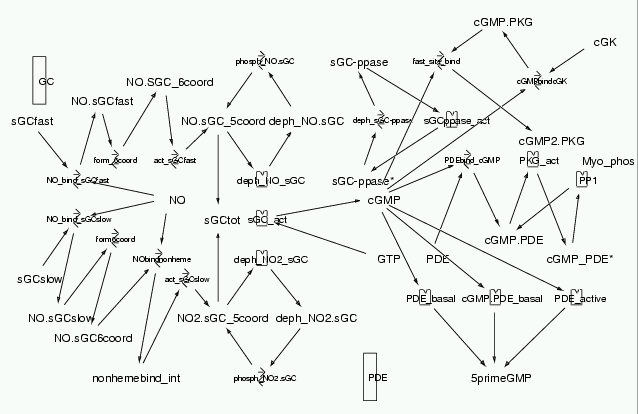
Pathway Basic Parameters | Pathway Name | Shared_Object_cGMP_regulation | | Pathway Entry Number | 176 | | Accession Number | 34 | | Accession Name | cGMP_regulation | | Species | Bovine and Sprague-Dawley rats; mammalian | | Tissue | Pulmonary, cardiac and expressed in Baculovirus infected Sf21(Spodoptera frugiperda) insect cells | | CellCompartment | Cytosol | | Related Pathways |
| | Notes | Though Corbin JD. et al. Eur J Biochem. (2000) 267(9):2760-7 has been mentioned in the citation, this model has been made with inputs from different literature sources, each of which has been mentioned in the notes sections. This model features hydrolysis of cGMP by bovine PDE, phosphorylation of PDE by bovine lung PKG, and activation of bovine lung PKG by cGMP binding. These mechanisms are known to be involved in cGMP level regulation. Rates have been used from different sources and the model has been tested based on Corbin JD. et al., since their work involved measuring the PDE phosphorylation and PDE activity.
On replicating Figures 2, 3 and 4 from their paper, there is approx 30% difference in results but the qualitative shape of the curves is very similar. This might be due to the fact that the Vmax values were used from different literature sources. This might lead to the discrepancy in the numbers in this model. The values shown in this model are near estimated physiological levels.In order to replicate the Figures more closely, we have run additional simulations with concentration terms changed so as to replicate the experimental conditions exactly.
| Statistics | Enzyme sites | Reactions | 1 | 0 | 0 |
Conversion formatThis pathway is part of accession 34 and is completely specified in the file acc34.g.
There is no separate files for just this pathway. |
| Format | File | | Native Format (GENESIS format) | acc34.g | | GENESIS Format (Annotated version) | Anno_acc34.g |
Pathway Detail Molecule List Enzyme List Reaction List
| Database compilation and code copyright (C) 2022, Upinder S. Bhalla and NCBS/TIFR
This Copyright is applied to ensure that the contents of this database remain freely available. Please see FAQ for details. |
|

