 |  |  GC GC |
Enter a Search String | | Special character and space not allowed in the query term.
Search string should be at least 2 characters long. |
Parameters for PDE (Pathway Number 178) Pathway Layout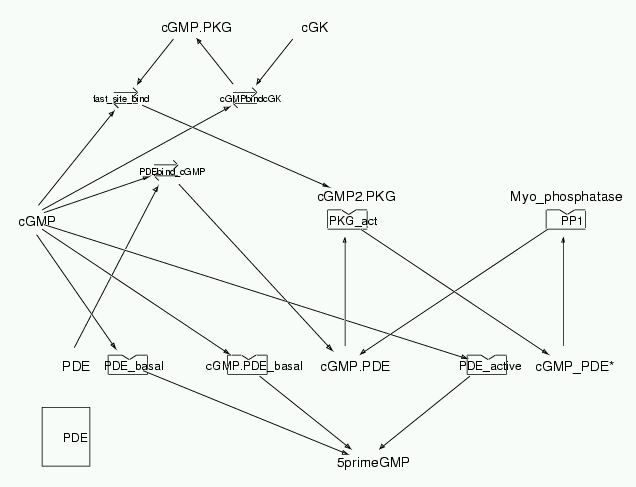
Pathway Basic Parameters | Pathway Name | PDE | | Pathway Entry Number | 178 | | Accession Number | 34 | | Accession Name | cGMP_regulation | | Species | Bovine and Sprague-Dawley rats; mammalian | | Tissue | Pulmonary, cardiac and expressed in Baculovirus infected Sf21(Spodoptera frugiperda) insect cells | | CellCompartment | Cytosol | | Related Pathways |
| | Notes | This model features the hydrolysis of cGMP by PDE. (The PDE taken into consideration is PDE 5, a hgihly cGMP specific PDE, and is considered to be the main enzyme responsible for terminating the action of cGMP generated following the release of NO from Nitrergic nerves, or from vascular endothelial cells. (Gibson, Eur J Biochem,2001,411:1-10). PDE is phosphorylated by PKG. (Corbin et al., Eur J Biochem,2000,267:2760-2767). And occupation of the allosteric binding sites on PDE by cGMP is necessary for phosphorylation by PKG. (Gibson, Eur J Biochem, 2001,411:1-10). Three plots in the paper by Corbin et al., were replicated with this model. Though the shape of the curves matched well, the numbers could not be matched due to the differences in Vmax values used from different literature sources apart from Corbin et al. Various inhibitors were used by Corbin et al., in their expts and so to match the numbers in the plots we have to replicate the experimental conditions exactly. So by bringing down the concentration of PKG and PDE we could match the numbers as well. But the values shown in this model are near physiological levels, as thats what we intend to do basically -- Building models of signaling pathways whose behaviour is similar to what happens invivo.
| Statistics Conversion formatThis pathway is part of accession 34 and is completely specified in the file acc34.g.
There is no separate files for just this pathway. |
| Format | File | | Native Format (GENESIS format) | acc34.g | | GENESIS Format (Annotated version) | Anno_acc34.g |
Pathway Detail Molecule List Enzyme List Reaction List
| Database compilation and code copyright (C) 2022, Upinder S. Bhalla and NCBS/TIFR
This Copyright is applied to ensure that the contents of this database remain freely available. Please see FAQ for details. |
|

