 |  |  PLA2 PLA2 |
Enter a Search String | | Special character and space not allowed in the query term.
Search string should be at least 2 characters long. |
Parameters for Ras (Pathway Number 58) Pathway Layout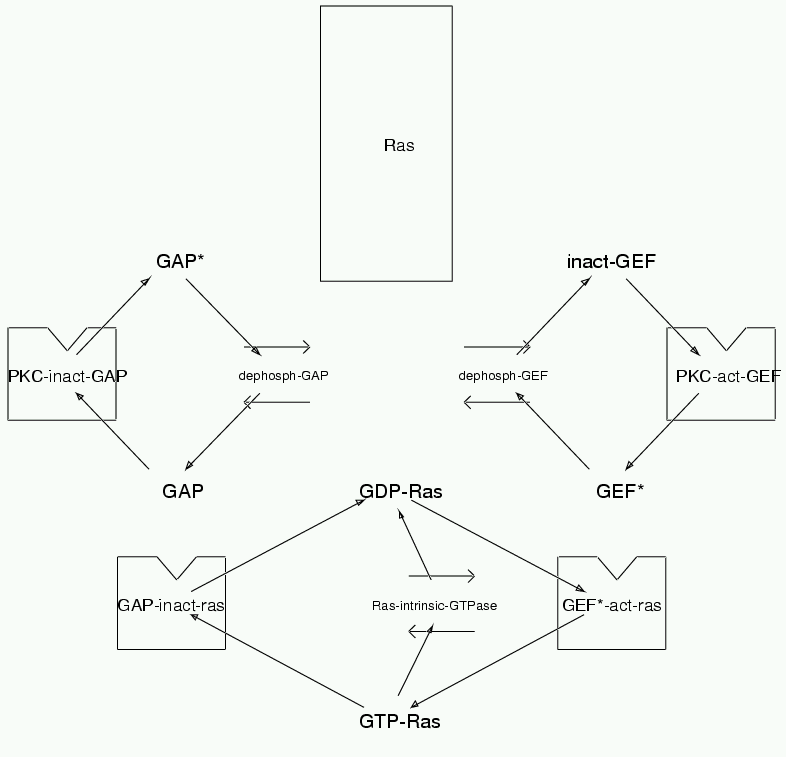
Pathway Basic Parameters | Pathway Name | Ras | | Pathway Entry Number | 58 | | Accession Number | 8 | | Accession Name | 3d_fold_model | | Species | Generic mammalian | | Tissue | NIH 3T3 Expression | | CellCompartment | Surface - Nucleus | | Related Pathways | 7,
25,
37,
63,
76,
184,
212,
231,
318,
333,
353,
367,
372,
378,
385,
391,
397,
404,
410,
416,
422,
427,
433,
439,
445,
451,
457,
463,
469,
475,
481,
487,
493,
499,
505,
511,
517,
523,
529,
536,
542,
548,
554,
560,
566,
572,
578,
584,
590,
596,
602,
608,
614,
620,
626,
632,
638,
644,
650,
656,
662,
668,
674,
680,
687,
693,
700,
705,
711,
717,
723,
729,
735,
741,
747,
753,
759,
765,
771,
777,
783,
789,
795,
801,
807,
813,
819,
825,
831,
837,
843,
849,
855,
861,
867,
873,
879,
885,
891,
897,
903,
909,
915,
923,
929,
935,
941,
947,
953,
959,
966,
972,
978,
984,
990,
996,
1001,
1007,
1013,
1019,
1025,
1031,
1037,
1043,
1049,
1055,
1061,
1067
| | Notes | Ras has now gotten to be a big enough component of the model to deserve its own group. The main refs are Boguski and McCormick Nature 366 643-654 '93 Major review Eccleston et al JBC 268:36 pp 27012-19 Orita et al JBC 268:34 2554246
| Statistics Conversion formatThis pathway is part of accession 8 and is completely specified in the file acc8.g.
There is no separate files for just this pathway. |
Pathway Detail Molecule List Enzyme List Reaction List
| Database compilation and code copyright (C) 2022, Upinder S. Bhalla and NCBS/TIFR
This Copyright is applied to ensure that the contents of this database remain freely available. Please see FAQ for details. |
|

