 |

|  CaRegulation CaRegulation |
Enter a Search String | | Special character and space not allowed in the query term.
Search string should be at least 2 characters long. |
Accession information for Osc_Ca_IP3metabolism (Accession Number 32)
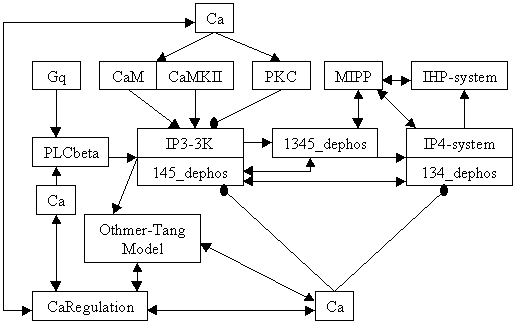
Reaction Scheme
Accession Basic Parameters
| Name | Osc_Ca_IP3metabolism |
| Accession Type | Network |
| Transcriber | Jyoti Mishra, NCBS |
| Developer | Jyoti Mishra, NCBS |
| Entry Date (YYYY-MM-DD) | 2002-04-03 00:00:00 |
| Species | Generic mammalian |
| Tissue | Brain - Neuronal |
| Cell Compartment | Cytosol |
| Source | Mishra J, Bhalla US. Biophys J. 2002 Sep;83(3):1298-316. ( peer-reviewed publication ) |
| Methodology | Quantitative match to experiments, Qualitative |
| Model Implementation | Exact GENESIS implementation |
| Model Validation | Approximates original data |
| Notes | This network models an oscillatory calcium response to GPCR mediated PLCbeta activation, alongwith detailed InsP3 metabolism in the neuron. It is similar to the Osc_Ca_IP3metab model (accession 24) except that some enzymes in the InsP3 metabolism network have been modified to have reversible kinetics rather than Michaelis-Menten kinetics. The modified enzymes belong to the groups: IP4-system, IP3-3K, 145_dephos and 134_dephos. Mishra J, Bhalla US. Biophys J. 2002 Sep;83(3):1298-316. |
Conversion formats| Format | File | | Native Format (GENESIS format) | acc32.g | | GENESIS Format (Annotated version) | Anno_acc32.g |
| Database compilation and code copyright (C) 2022, Upinder S. Bhalla and NCBS/TIFR
This Copyright is applied to ensure that the contents of this database remain freely available. Please see FAQ for details. |
|

