 |  |  CaRegulation CaRegulation |
Enter a Search String | | Special character and space not allowed in the query term.
Search string should be at least 2 characters long. |
Parameters for Othmer-Tang-model (Pathway Number 171) Pathway Layout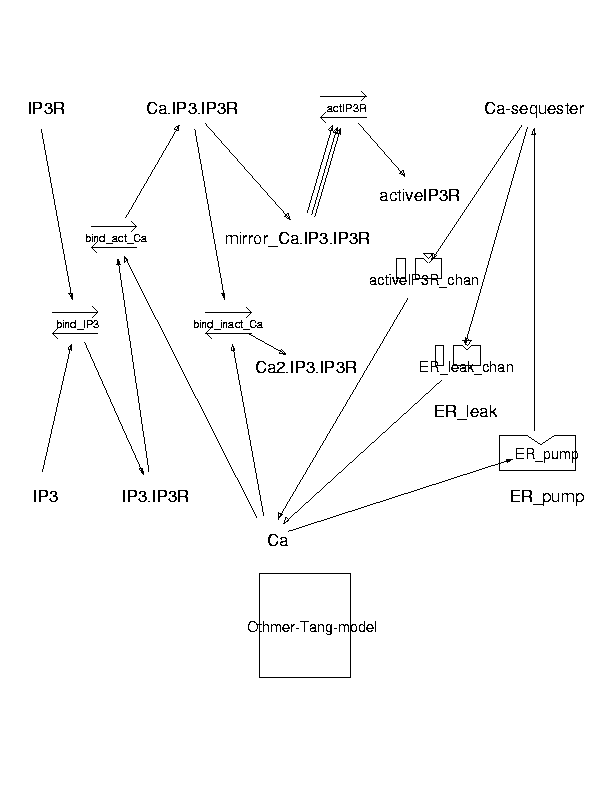
Pathway Basic Parameters | Pathway Name | Othmer-Tang-model | | Pathway Entry Number | 171 | | Accession Number | 32 | | Accession Name | Osc_Ca_IP3metabolism | | Species | Generic mammalian | | Tissue | Brain - Neuronal | | CellCompartment | Cytosol | | Related Pathways | 133
| | Notes | The Othmer-Tang kinetic model for the InsP3 receptor with oscillatory Ca dynamics. This model incorporates both positive and negative feedback from calcium. The active IP3 receptor channel has one Ca and one IP3 bound. The inactive state of the channel (post-activation) is represented by 2 Ca and one IP3 bound to the IP3 receptor. Parameters in this model are as far as possible identical to those used by Othmer and Tang (Tang et al, Biophys J 70; 1996: 246-63).
| Statistics Conversion formatThis pathway is part of accession 32 and is completely specified in the file acc32.g.
There is no separate files for just this pathway. |
| Format | File | | Native Format (GENESIS format) | acc32.g | | GENESIS Format (Annotated version) | Anno_acc32.g |
Pathway Detail Molecule List Enzyme List Reaction List
| Database compilation and code copyright (C) 2022, Upinder S. Bhalla and NCBS/TIFR
This Copyright is applied to ensure that the contents of this database remain freely available. Please see FAQ for details. |
|

