
|
Enter a Search String | | Special character and space not allowed in the query term.
Search string should be at least 2 characters long. |
Parameters for RasGTPase (Pathway Number 307) Pathway Layout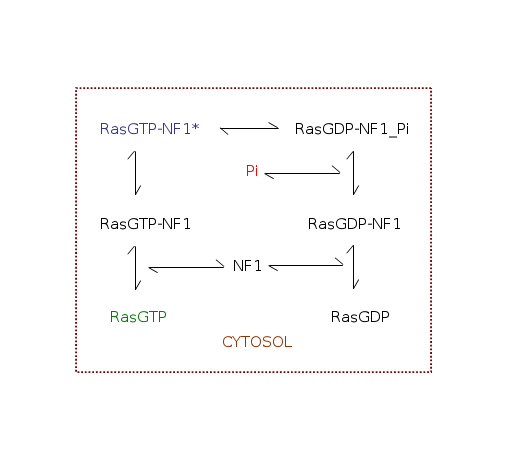
Pathway Basic Parameters | Pathway Name | RasGTPase | | Pathway Entry Number | 307 | | Accession Number | 71 | | Accession Name | RasGTPase | | Species | General Mammalian | | Tissue | E.coli Expression system | | CellCompartment | Cytosol | | Related Pathways |
| | Notes | Ras is an important regulator of cell growth in all eukaryotic cells. The model represent hydrolysis of active Ras-bound GTP to give inactive Ras-bound GDP catalyzed by GTPase activating proteins i.e NF1. The inactive Ras-bound GDP turns signalling protein off.
| Statistics Conversion formatThis pathway is part of accession 71 and is completely specified in the file acc71.g.
There is no separate files for just this pathway. |
Pathway Detail Molecule List Enzyme List Reaction List
| Database compilation and code copyright (C) 2022, Upinder S. Bhalla and NCBS/TIFR
This Copyright is applied to ensure that the contents of this database remain freely available. Please see FAQ for details. |
|

