|
Enter a Search String | | Special character and space not allowed in the query term.
Search string should be at least 2 characters long. |
Parameters for L-Arginine (Pathway Number 308) Pathway Layout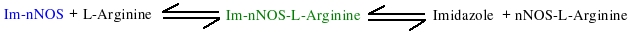
Pathway Basic Parameters | Pathway Name | L-Arginine | | Pathway Entry Number | 308 | | Accession Number | 72 | | Accession Name | L-Arginine | | Species | Human | | Tissue | Kidney cell expressing the rat brain nNOS | | CellCompartment | Cytosol | | Related Pathways |
| | Notes | This model was taken from the Abu-Soud HM et al. Biochemistry. 1999 Sep 21;38(38):12446-51. This model shows kinetic binding of L-Arginine to neuronal nitric oxide synthase.
Model shows the substrate (L-Arginine) binding to nNOS in a two-step reversible fashion. First there is rapid binding equilibrium between Im-nNOS and L-Arginine to form an intermediate that contains bound imidazole and L-Arginine. This is followed by a slower conformational change in the Im-enzyme-substrate complex that is associated with release of bound imidazole and generation of a modified enzyme-substrate complex which is detected due to spectral change. Replicates Figure 4 (right panel).
| Statistics Conversion formatThis pathway is part of accession 72 and is completely specified in the file acc72.g.
There is no separate files for just this pathway. |
Pathway Detail Molecule List Enzyme List Reaction List
| Database compilation and code copyright (C) 2022, Upinder S. Bhalla and NCBS/TIFR
This Copyright is applied to ensure that the contents of this database remain freely available. Please see FAQ for details. |
|