 |  |  IHP-system IHP-system |
Enter a Search String | | Special character and space not allowed in the query term.
Search string should be at least 2 characters long. |
Parameters for CaMKII (Pathway Number 106) Pathway Layout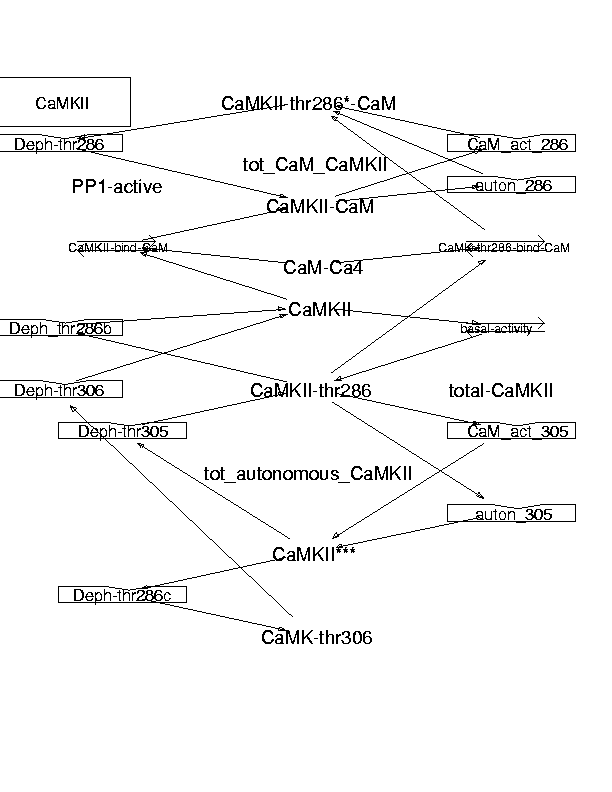
Pathway Basic Parameters | Pathway Name | CaMKII | | Pathway Entry Number | 106 | | Accession Number | 23 | | Accession Name | NonOsc_Ca_IP3metabolism | | Species | Generic mammalian | | Tissue | Brain - Neuronal | | CellCompartment | Cytosol | | Related Pathways | 13,
26,
80,
121,
145,
159,
174,
202,
216,
235,
245,
258,
264,
272,
282,
322,
339,
357
| | Notes | Main reference here is the review by Hanson PI, Schulman H. Annu Rev Biochem. 1992;61:559-601. Most of the mechanistic details and a few constants are derived from there. Many kinetics are from Hanson PI, Schulman H. J Biol Chem. 1992 Aug 25;267(24):17216-24. The enzymes look a bit complicated. Actually it is just 3 reactions for different sites, by 4 states of CaMKII, defined by the phosphorylation state. This model approximates the fact that the enzyme is actually present as a decamer/dodecamer. It does so by treating the autophosphorylation reactions as being independent of the concentration of CaMKII. Also the rates for the autophosphorylation steps have been scaled to fit this approximation.
| Statistics Conversion formatThis pathway is part of accession 23 and is completely specified in the file acc23.g.
There is no separate files for just this pathway. |
| Format | File | | Native Format (GENESIS format) | acc23.g | | GENESIS Format (Annotated version) | Anno_acc23.g |
Pathway Detail Molecule List Enzyme List Reaction List
| Database compilation and code copyright (C) 2022, Upinder S. Bhalla and NCBS/TIFR
This Copyright is applied to ensure that the contents of this database remain freely available. Please see FAQ for details. |
|

