 |  |  CaRegulation CaRegulation |
Enter a Search String | | Special character and space not allowed in the query term.
Search string should be at least 2 characters long. |
Parameters for 145_dephos (Pathway Number 166) Pathway Layout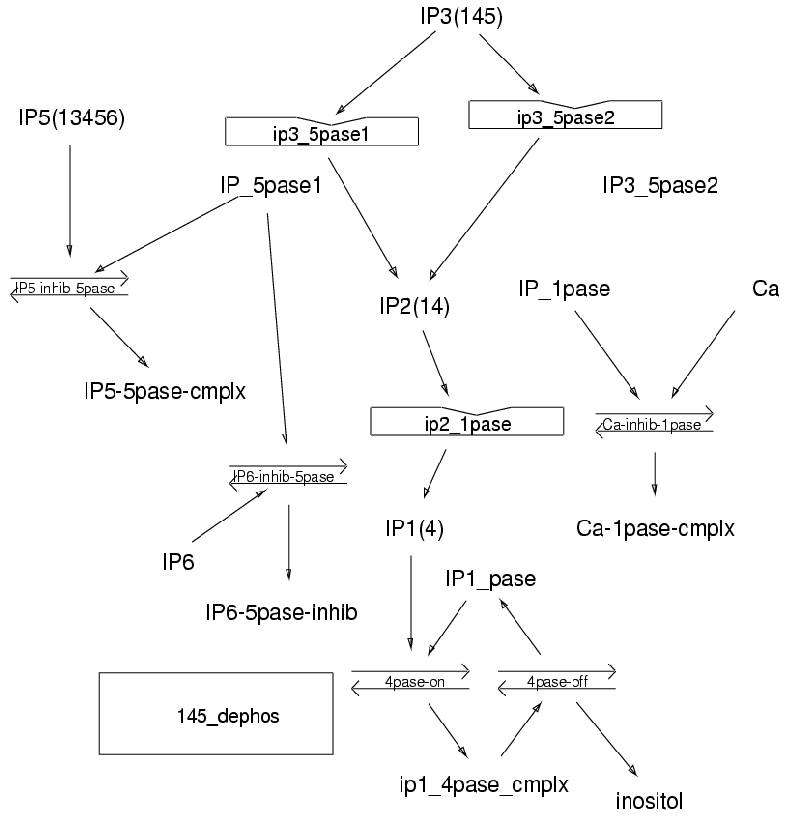
Pathway Basic Parameters | Pathway Name | 145_dephos | | Pathway Entry Number | 166 | | Accession Number | 32 | | Accession Name | Osc_Ca_IP3metabolism | | Species | Generic mammalian | | Tissue | Brain - Neuronal | | CellCompartment | Cytosol | | Related Pathways | 114,
128,
153
| | Notes | Model for Ins(145)P3 dephosphorylation. InsP3 degrades via Ins(14)P2 and Ins(4)P2 to inositol. Model incorporates enzyme inhibition from higher phosphates: InsP5 and InsP6. Primary refs: Hansen et al, JBC 262, 1987: 17319-26; Hoer and Oberdisse, BiochemJ 278, 1991: 219-24; Inhorn and Majerus, JBC 262, 1987: 15946-52; Gee et al, BiochemJ 249, 1988: 883-89
| Statistics Conversion formatThis pathway is part of accession 32 and is completely specified in the file acc32.g.
There is no separate files for just this pathway. |
| Format | File | | Native Format (GENESIS format) | acc32.g | | GENESIS Format (Annotated version) | Anno_acc32.g |
Pathway Detail Molecule List Enzyme List Reaction List
| Database compilation and code copyright (C) 2022, Upinder S. Bhalla and NCBS/TIFR
This Copyright is applied to ensure that the contents of this database remain freely available. Please see FAQ for details. |
|

