 |  |  Ras Ras |
Enter a Search String | | Special character and space not allowed in the query term.
Search string should be at least 2 characters long. |
Parameters for PLA2 (Pathway Number 183) Pathway Layout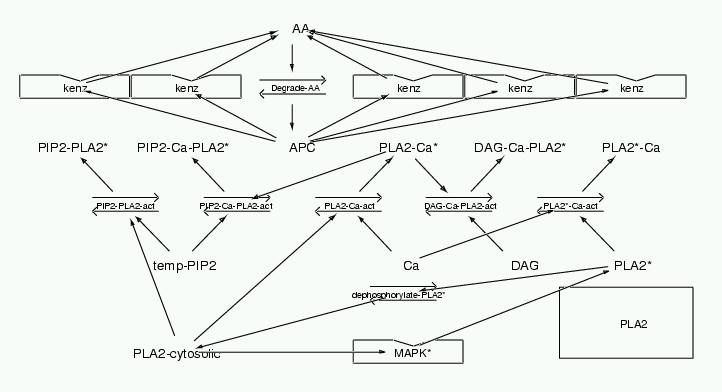
Pathway Basic Parameters | Pathway Name | PLA2 | | Pathway Entry Number | 183 | | Accession Number | 35 | | Accession Name | MAPK-bistability-fig1c | | Species | Generic mammalian | | Tissue | NIH 3T3 Expression | | CellCompartment | Surface - Nucleus | | Related Pathways | 3,
21,
36,
57,
62,
72,
208,
314,
331,
349,
365,
920,
922,
933,
939,
945,
951,
957,
964,
970,
976,
982,
988,
994,
1000,
1005,
1011,
1017,
1023,
1029,
1035,
1041,
1047,
1053,
1059,
1065
| | Notes | Main source of data: Leslie and Channon BBA 1045 (1990) pp 261-270. Fig 6 is Ca curve. Fig 4a is PIP2 curve. Fig 4b is DAG curve. Also see Wijkander and Sundler JBC 202 (1991) pp873-880; Diez and Mong JBC 265(24) p14654; Leslie JBC 266(17) (1991) pp11366-11371 Many inputs activate PLA2. In this model I simply take each combination of stimuli as binding to PLA2 to give a unique enzymatic activity. The Km and Vmax of these active complexes is scaled according to the relative activation reported in the papers above.
| Statistics Conversion formatThis pathway is part of accession 35 and is completely specified in the file acc35.g.
There is no separate files for just this pathway. |
Pathway Detail Molecule List Enzyme List Reaction List
| Database compilation and code copyright (C) 2022, Upinder S. Bhalla and NCBS/TIFR
This Copyright is applied to ensure that the contents of this database remain freely available. Please see FAQ for details. |
|

