 |  |  AMPAR AMPAR |
Enter a Search String | | Special character and space not allowed in the query term.
Search string should be at least 2 characters long. |
Parameters for CaM (Pathway Number 236) Pathway Layout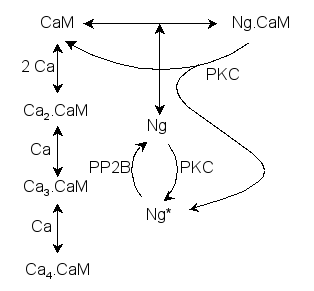
Pathway Basic Parameters | Pathway Name | CaM | | Pathway Entry Number | 236 | | Accession Number | 59 | | Accession Name | AMPAR_traff_model0 | | Species | General Mammalian | | Tissue | Neuronal | | CellCompartment | Synaptic Spine, Postsynaptic Density | | Related Pathways | 14,
27,
81,
107,
122,
146,
160,
173,
203,
217,
246,
259,
265,
273,
283,
323,
340,
358,
368,
373,
379,
386,
392,
398,
405,
411,
418,
423,
428,
434,
440,
446,
452,
458,
464,
470,
476,
482,
488,
494,
500,
506,
512,
518,
524,
530,
537,
543,
549,
555,
561,
568,
573,
579,
585,
591,
597,
603,
609,
615,
621,
627,
633,
639,
645,
651,
657,
663,
669,
675,
681,
688,
695,
701,
706,
712,
718,
724,
730,
736,
742,
748,
754,
760,
766,
772,
778,
784,
790,
796,
802,
808,
814,
820,
826,
832,
838,
844,
850,
856,
862,
868,
874,
880,
886,
892,
898,
904,
910,
916,
924,
930,
936,
942,
948,
954,
961,
967,
973,
979,
985,
991,
997,
1002,
1008,
1014,
1021,
1026,
1032,
1038,
1044,
1050,
1056,
1062,
1068,
1094
| | Notes | This is model 0 from Hayer and Bhalla, PLoS Comput Biol 2005. It has a bistable model of AMPAR traffick, plus a
non-bistable model of CaMKII. This differs from the reference model (model 1) in that model0 lacks degradation and turno
ver reactions for AMPAR.
| Statistics Conversion formatThis pathway is part of accession 59 and is completely specified in the file acc59.g.
There is no separate files for just this pathway. |
Pathway Detail Molecule List Enzyme List Reaction List
| Database compilation and code copyright (C) 2022, Upinder S. Bhalla and NCBS/TIFR
This Copyright is applied to ensure that the contents of this database remain freely available. Please see FAQ for details. |
|

