 |  |  AC AC |
Enter a Search String | | Special character and space not allowed in the query term.
Search string should be at least 2 characters long. |
Parameters for PP1 (Pathway Number 266) Pathway Layout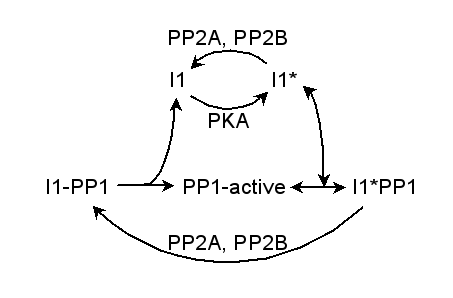
Pathway Basic Parameters | Pathway Name | PP1 | | Pathway Entry Number | 266 | | Accession Number | 63 | | Accession Name | CaMKII_model3 | | Species | General Mammalian | | Tissue | Neuronal | | CellCompartment | Synaptic Spine, Postsynaptic Density | | Related Pathways | 15,
28,
82,
204,
218,
237,
247,
260,
274,
284,
324,
341,
359
| | Notes | This is the complete model of CaMKII bistability, model 3. It exhibits bistability in CaMKII activation due to autophosphorylation at the PSD and local saturation of PP1. This version of model 3 includes PKA regulatory input. This has little effect on the deterministic calculations, but the PKA pathway introduces a lot of noise which causes a difference in stochastic runs.
| Statistics Conversion formatThis pathway is part of accession 63 and is completely specified in the file acc63.g.
There is no separate files for just this pathway. |
Pathway Detail Molecule List Enzyme List Reaction List
| Database compilation and code copyright (C) 2022, Upinder S. Bhalla and NCBS/TIFR
This Copyright is applied to ensure that the contents of this database remain freely available. Please see FAQ for details. |
|

