 |  |  Ras Ras |
Enter a Search String | | Special character and space not allowed in the query term.
Search string should be at least 2 characters long. |
Parameters for PKC (Pathway Number 34) Pathway Layout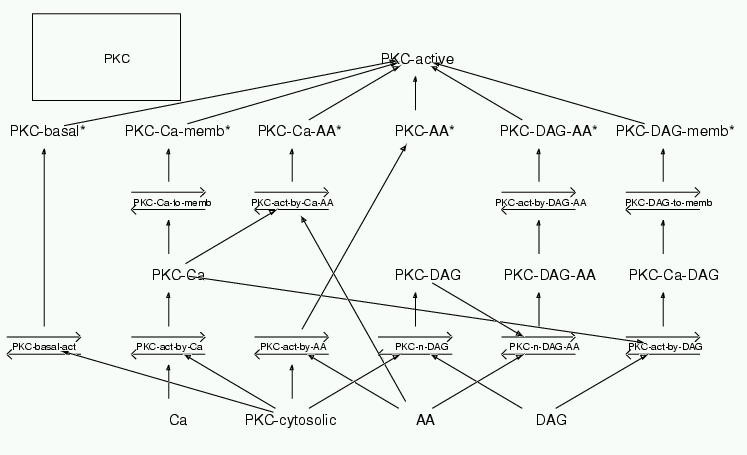
Pathway Basic Parameters | Pathway Name | PKC | | Pathway Entry Number | 34 | | Accession Number | 4 | | Accession Name | mkp1_feedback_effects | | Species | Generic mammalian | | Tissue | NIH 3T3 Expression | | CellCompartment | Surface - nucleus | | Related Pathways | 2,
20,
55,
60,
71,
108,
123,
138,
147,
161,
181,
200,
207,
313,
330,
348,
364,
370,
376,
383,
389,
395,
401,
408,
414,
420,
426,
431,
437,
443,
449,
455,
461,
467,
473,
479,
485,
491,
497,
503,
509,
515,
521,
527,
534,
540,
546,
552,
558,
564,
570,
576,
582,
588,
594,
600,
606,
612,
618,
624,
630,
636,
642,
648,
654,
660,
666,
672,
678,
685,
691,
698,
703,
709,
715,
721,
727,
733,
739,
745,
751,
757,
763,
769,
775,
781,
787,
793,
799,
805,
811,
817,
823,
829,
835,
841,
847,
853,
859,
865,
871,
877,
883,
889,
895,
901,
907,
913,
919,
927,
932,
938,
944,
950,
956,
963,
969,
974,
981,
987,
993,
999,
1004,
1010,
1016,
1022,
1028,
1034,
1040,
1046,
1052,
1058,
1064
| | Notes | Protein Kinase C. This module represents a weighted average of the alpha, beta and gamma isoforms. It takes inputs from Ca, DAG (Diacyl Glycerol) and AA (arachidonic acid). Regulation parameters are largely from Schaechter and Benowitz 1993 J Neurosci 13(10):4361 who use synaptosomes from mammalian brain and in one paper look at all three inputs. Shinomura et al 1991 PNAS 88:5149-5153 is also a useful source of data and helps to tighten the DAG inputs. General reviews include Azzi et al 1992 Eur J Bioch 208:541 and Nishizuka 1988, Nature 334:661 Concentration info from Kikkawa et al 1982 JBC 257(22):13341 The process of parameterization is described in detail in several places. See Supplementary notes to Bhalla and Iyengar 1999 Science 284:92-96, available at the site http://www.ncbs.res.in/~bhalla/ltploop/pkc_example.html The parameterization is also described in a book chapter: Bhalla, 2000: Simulations of Biochemical Signaling in Computational Neuroscience: Realistic Modeling for Experimentalists. Ed. E. De Schutter. CRC Press.
| Statistics Conversion formatThis pathway is part of accession 4 and is completely specified in the file acc4.g.
There is no separate files for just this pathway. |
Pathway Detail Molecule List Enzyme List Reaction List
| Database compilation and code copyright (C) 2022, Upinder S. Bhalla and NCBS/TIFR
This Copyright is applied to ensure that the contents of this database remain freely available. Please see FAQ for details. |
|

