 |  |  PKA PKA |
Enter a Search String | | Special character and space not allowed in the query term.
Search string should be at least 2 characters long. |
Parameters for MAPK (Pathway Number 352) Pathway Layout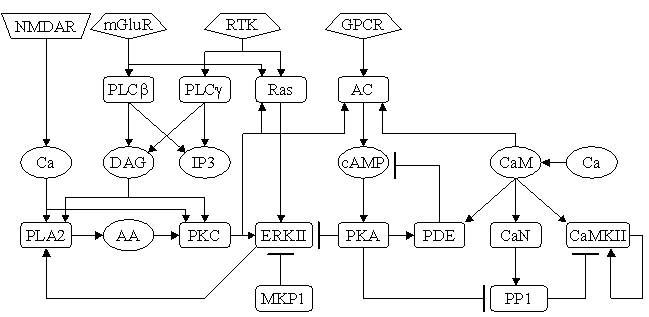
Pathway Basic Parameters | Pathway Name | MAPK | | Pathway Entry Number | 352 | | Accession Number | 78 | | Accession Name | Ajay_Bhalla_2004_Feedback_Tuning | | Species | Rat | | Tissue | Hippocampal CA1 | | CellCompartment | Dendrites | | Related Pathways | 6,
24,
35,
56,
61,
75,
139,
142,
182,
193,
211,
230,
317,
335,
366,
371,
377,
384,
390,
396,
402,
403,
409,
415,
421,
432,
438,
444,
450,
456,
462,
468,
474,
480,
486,
492,
498,
504,
510,
516,
522,
528,
535,
541,
547,
553,
559,
565,
571,
577,
583,
589,
595,
601,
607,
613,
619,
625,
631,
637,
643,
649,
655,
661,
667,
673,
679,
686,
692,
699,
704,
710,
716,
722,
728,
734,
740,
746,
752,
758,
764,
770,
776,
782,
788,
794,
800,
806,
812,
818,
824,
830,
836,
842,
848,
854,
860,
866,
872,
878,
884,
890,
896,
902,
908,
914,
921,
928,
934,
940,
946,
952,
958,
960,
965,
971,
977,
983,
989,
995,
1006,
1012,
1018,
1024,
1030,
1036,
1042,
1048,
1054,
1060,
1066
| | Notes | This model is taken from Ajay SM, Bhalla US. Eur J Neurosci. 2004 Nov;20(10):2671-80. This is the feedback model from Figure 8a.
| Statistics Conversion formatThis pathway is part of accession 78 and is completely specified in the file acc78.g.
There is no separate files for just this pathway. |
Pathway Detail Molecule List Enzyme List Reaction List
| Database compilation and code copyright (C) 2022, Upinder S. Bhalla and NCBS/TIFR
This Copyright is applied to ensure that the contents of this database remain freely available. Please see FAQ for details. |
|

