
|
Enter a Search String | | Special character and space not allowed in the query term.
Search string should be at least 2 characters long. |
Parameters for MAPK (Pathway Number 139) Pathway Layout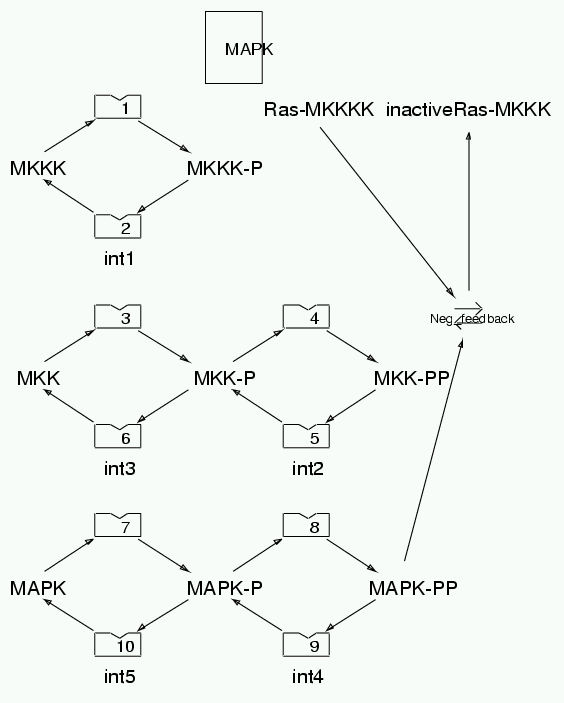
Pathway Basic Parameters | Pathway Name | MAPK | | Pathway Entry Number | 139 | | Accession Number | 27 | | Accession Name | MAPK_osc | | Species | Xenopus | | Tissue | Oocyte extract | | CellCompartment | Cytosol | | Related Pathways | 6,
24,
35,
56,
61,
75,
142,
182,
193,
211,
230,
317,
335,
352,
366,
371,
377,
384,
390,
396,
402,
403,
409,
415,
421,
432,
438,
444,
450,
456,
462,
468,
474,
480,
486,
492,
498,
504,
510,
516,
522,
528,
535,
541,
547,
553,
559,
565,
571,
577,
583,
589,
595,
601,
607,
613,
619,
625,
631,
637,
643,
649,
655,
661,
667,
673,
679,
686,
692,
699,
704,
710,
716,
722,
728,
734,
740,
746,
752,
758,
764,
770,
776,
782,
788,
794,
800,
806,
812,
818,
824,
830,
836,
842,
848,
854,
860,
866,
872,
878,
884,
890,
896,
902,
908,
914,
921,
928,
934,
940,
946,
952,
958,
960,
965,
971,
977,
983,
989,
995,
1006,
1012,
1018,
1024,
1030,
1036,
1042,
1048,
1054,
1060,
1066
| | Notes | This is the oscillatory MAPK model from Kholodenko 2000 Eur J. Biochem 267:1583-1588 The original model is formulated in terms of idealized Michaelis-Menten enzymes and the enzyme-substrate complex concentrations are therefore assumed negligible. The current implementation of the model uses explicit enzyme reactions involving substrates and is therefore an approximation to the Kholodenko model. The approximation is greatly improved if the enzyme is flagged as Available which is an option in Kinetikit. This flag means that the enzyme protein concentration is not reduced even when it is involved in a complex. However, the substrate protein continues to participate in enzyme-substrate complexes and its concentration is therefore affected. Overall, this model works almost the same as the Kholodenko model but the peak MAPK-PP amplitudes are a little reduced and the period of oscillations is about 10% longer. If the enzymes are not flagged as Available then the oscillations commence only when the Km for enzyme 1 is set to 0.1 uM.
| Statistics Conversion formatThis pathway is part of accession 27 and is completely specified in the file acc27.g.
There is no separate files for just this pathway. |
Pathway Detail Molecule List Enzyme List Reaction List
| Database compilation and code copyright (C) 2022, Upinder S. Bhalla and NCBS/TIFR
This Copyright is applied to ensure that the contents of this database remain freely available. Please see FAQ for details. |
|

